Overview of the project
In this project, we develop a system called CLAP that detects and classifies “potentially unwanted applications” (PUAs) such as adware or remote monitoring tools. The number of PUAs that trigger alerts in network security systems has increased to a point where their analysis overwhelms the capacity of incident response teams. Since the team must focus on the malware that presents more critical threats, there is a growing demand for systems that can systematically distinguish PUAs from malware and benign applications. Our approach leverages DNS queries made by apps. Using a large sample of Android apps from third-party marketplaces, we first reveal that DNS queries can provide useful information for detection and classification of PUAs. We have validated that existing DNS blacklists are limited when performing these tasks and have clarified that the CLAP system performs with high accuracy.
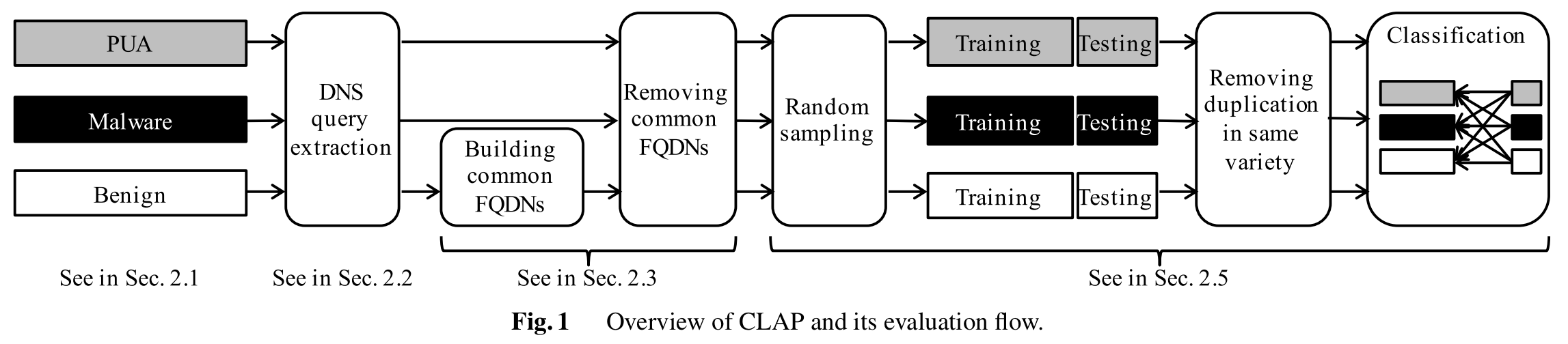
Dataset
Herein, we provide the dataset we used in this research project:
The dataset includes 5,340 PUAs that account for 237 of distinct varieties. PUAs were identified by searching for certain keywords (“pua”, “pup”, “adware”, “unwanted”, “ ad”, and “/ad”) in the detection name by anti-virus software. The dataset also includes 5,340 malware and benign apps. The fields of csv are label, detection name by anti-virus software, sha1sum, app market, file name, and extracted FQDNs.
If you are interested in accessing to the CLAP dataset, please send us an email from your university’s or company’s email account.
Email: clap@nsl.cs.waseda.ac.jp
In your email, please include your name, affiliation, and your purpose to use the dataset. We use the information for verification. If you are a student, please indicate the name of your supervisor and her/his affiliation.
If your papers or articles use our dataset, please cite our paper below.
Publication
- Mitsuhiro HATADA and Tatsuya MORI, “CLAP: Classification of Android PUAs by Similarity of DNS Queries,” IEICE TRANSACTIONS on Information and Systems, Vol. 103-D, No. 2, pp. 265–275, January 2020 [PDF]
Acknowledgements
A part of this work was supported by JSPS Grant-in-Aid for Scientific Research B, Grant Number JP16H02832.
